Enhancing the stability of DNA origami nanostructures: staple strand redesign versus enzymatic ligation†
Abstract
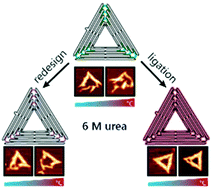
DNA origami structures have developed into versatile tools in molecular sciences and nanotechnology. Currently, however, many potential applications are hindered by their poor stability, especially under denaturing conditions. Here we present and evaluate two simple approaches to enhance DNA origami stability. In the first approach, we elevated the melting temperature of nine critical staple strands by merging the oligonucleotides with adjacent sequences. In the second approach, we increased the global stability by enzymatically ligating all accessible staple strand ends directly. By monitoring the gradual urea-induced denaturation of a prototype triangular DNA origami modified by these approaches using atomic force microscopy, we show that rational redesign of a few, critical staple strands leads to a considerable increase in overall stability at high denaturant concentration and elevated temperatures. In addition, enzymatic ligation yields DNA nanostructures with superior stability at up to 37 °C and in the presence of 6 M urea without impairing their shape. This bio-orthogonal approach is readily adaptable to other DNA origami structures without the need for synthetic nucleotide modifications when structural integrity under harsh conditions is required.


 Please wait while we load your content...
Please wait while we load your content...