Defect-assisted protein HP35 denaturation on graphene†
Abstract
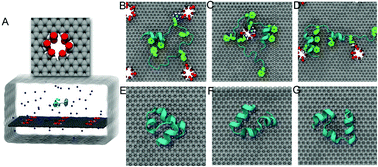
Structural defects in nanomaterials can alter their physical and chemical properties including magnetization, electronic and thermal conductivities, light absorption, and emission capabilities. Here, we investigated the potential impact of these defects on their biological effects through molecular dynamics simulations. By modeling the interaction between a graphene nanosheet and a widely used model protein, the chicken villin headpiece subdomain (HP35), we observed severe protein denaturation upon contact with defective graphene, while the protein remained intact on ideal graphene. The enhanced toxicity of defective graphene was due to the stronger attraction of the surface residues of HP35 from the defect edges (represented by carboxyl groups in our simulations) than from the ideal graphene. Upon binding to defective graphene, the contacting residues were restrained near the defective sites, which acted as “anchors” for the adsorbed protein. The “anchors” subsequently caused the protein to expose its aromatic and hydrophobic core residues to the graphene surface, via strong π–π stacking and hydrophobic interactions, thus leading to the unfolding of the protein. These findings not only highlight the importance of defects in nanomaterials’ impact on biological systems, but also provide insights into fine-tuning the potential biological properties of nanomaterials through defect engineering.
- This article is part of the themed collection: Nanoscale 10th Anniversary Special Issue


 Please wait while we load your content...
Please wait while we load your content...