Employing LiCl salt gradient in the wild-type α-hemolysin nanopore to slow down DNA translocation and detect methylated cytosine†
Abstract
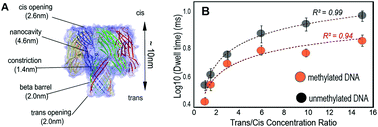
In this research, we demonstrate a label-free detection, biological nanopore-based method to distinguish methylated cytosine (mC) from naked cytosine (C) in sample mixtures containing both C and mC at a prolonged translocation duration. Using a 15-fold increase in LiCl salt concentration going from a cis to trans chamber, we increased the translocation dwell time of ssDNA by over 5-fold and the event capture rate by 6-fold in comparison with the symmetric concentration of 1.0 M KCl (control). This is a consequence of counter-ion binding and effective lowering of the overall charge of DNA, which in turn lessens the electrophoretic drive of the system and slows the translocation velocity. Moreover, salt gradients can create a large electric field that will funnel ions and polymers towards the pore, increasing the capture rate and translocation dwell time of DNA. As a result, in 0.2 M–3.0 M LiCl solution, ssDNA achieved a prolonged dwell time of 52 μs per nucleotide and a capture rate of 60 ssDNA per second. Importantly, lowering the translocation speed of ssDNA enhances the resulting resolution, allowing 5′-mC to be distinguished from C without using methyl-specific labels. We successfully distinguished 5′-mC from C when mixed together at ratios of 1 : 1, 3 : 7 and 7 : 3. The distribution of current blockade amplitudes of all mixtures adopted bimodal shapes, with peak-to-peak ratios coarsely corresponding to the mixture composition (e.g. the density and distribution of events shifted in correspondence with changes in 18b-0mC and 18-2mC concentration ratios in the mixture).


 Please wait while we load your content...
Please wait while we load your content...