Prediction of S-nitrosylation sites by integrating support vector machines and random forest†
Abstract
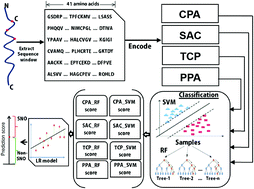
Cysteine S-nitrosylation is a type of reversible post-translational modification of proteins, which controls diverse biological processes. It is associated with redox-based cellular signaling to protect against oxidative stress. The identification of S-nitrosylation sites is an important step to reveal the function of proteins; however, experimental identification of S-nitrosylation is expensive and time-consuming work. Hence, sequence-based computational prediction of potential S-nitrosylation sites is highly sought before experimentation. Herein, a novel predictor PreSNO has been developed that integrates multiple encoding schemes by the support vector machine and random forest algorithms. The PreSNO achieved an accuracy and Matthews correlation coefficient value of 0.752 and 0.252 respectively in classifying between SNO and non-SNO sites when evaluated on the independent dataset, outperforming the existing methods. The web application of the PreSNO and its associated datasets are freely available at http://kurata14.bio.kyutech.ac.jp/PreSNO/.


 Please wait while we load your content...
Please wait while we load your content...