Prediction of zinc-binding sites using multiple sequence profiles and machine learning methods†
Abstract
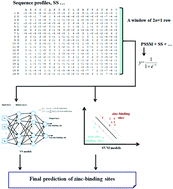
The zinc (Zn2+) cofactor has been proven to be involved in numerous biological mechanisms and the zinc-binding site is recognized as one of the most important post-translation modifications in proteins. Therefore, accurate knowledge of zinc ions in protein structures can provide potential clues for elucidation of protein folding and functions. However, determining zinc-binding residues by experimental means is usually lab-intensive and associated with high cost in most cases. In this context, the development of computational tools for identifying zinc-binding sites is highly desired, especially in the current post-genomic era. In this work, we developed a novel zinc-binding site prediction method by combining several intensively-trained machine learning models. To establish an accurate and generative method, we downloaded all zinc-binding proteins from the Protein Data Bank and prepared a non-redundant dataset. Meanwhile, a well-prepared dataset by other groups was also used. Then, effective and complementary features were extracted from sequences and three-dimensional structures of these proteins. Moreover, several well-designed machine learning models were intensively trained to construct accurate models. To assess the performance, the obtained predictors were stringently benchmarked using the diverse zinc-binding sites. Furthermore, several state-of-the-art in silico methods developed specifically for zinc-binding sites were also evaluated and compared. The results confirmed that our method is very competitive in real world applications and could become a complementary tool to wet lab experiments. To facilitate research in the community, a web server and stand-alone program implementing our method were constructed and are publicly available at http://bioinformatics.fzu.edu.cn/znMachine.html. The downloadable program of our method can be easily used for the high-throughput screening of potential zinc-binding sites across proteomes.


 Please wait while we load your content...
Please wait while we load your content...