Parallelized identification of on- and off-target protein interactions†
Abstract
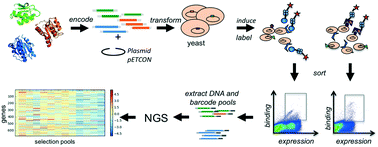
Genetic selection combined with next-generation sequencing enables the simultaneous interrogation of the functionality and stability of large numbers of naturally occurring, engineered, or computationally designed protein variants in parallel. We display hundreds of engineered proteins on the surface of yeast cells, select for binding to a set of target molecules by flow cytometry, and sequence the starting pool as well as selected pools to obtain enrichment values for each displayed protein with each target. We show that this high-throughput workflow of multiplex genetic selections followed by large-scale sequencing and comparative analysis allows not only the determination of relative affinities, but also the monitoring of specificity profiles for hundreds to thousands of protein–protein and protein–small molecule interactions in parallel. The approach not only identifies new interactions of designed proteins, but also detects unintended and undesirable off-target interactions. This provides a general framework for screening of engineered protein binders, which often have no negative selection or design step as part of their development pipelines. Hence, this method will be generally useful in the development of protein-based therapeutics.
- This article is part of the themed collection: MSDE Emerging Investigators 2020


 Please wait while we load your content...
Please wait while we load your content...
