A microfluidic alternating-pull–push active digitization method for sample-loss-free digital PCR†
Abstract
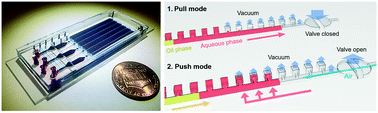
Digital polymerase chain reaction (dPCR) is a powerful tool for genetic analysis, providing superior sensitivity and accuracy. In many applications that demand minuscule reaction volumes, such as single cell analysis, efficient and reproducible sample handling and digitization is pivotal for accurate absolute quantification of targets, but remains a significant technical challenge. In this paper, we described a robust and flexible microfluidic alternating-pull–push active digitization (μAPPAD) strategy that confers close to 100% sample digitization efficiency for microwell-based dPCR. Our strategy employs pneumatic valve control to periodically manipulate air pressure inside the chip to greatly facilitate the vacuum-driven partition of solution into microwells, enabling efficient digitization of a small-volume solution with significantly reduced volume variability. The μAPPAD method was evaluated on both tandem-channel and parallel-channel chips, which achieved a digitization efficiency of 99.5 ± 0.3% and 94.6 ± 0.9% within 10.5 min and 2 min, respectively. To assess the analytical performance of the μAPPAD chip, we calibrated it for absolution dPCR quantitation of λDNA across a range of concentrations. The results obtained with our chip matched well with the theoretical curve computed from Poisson statistics. Compared to the existing methods for highly efficient sample digitization, not only does our technology greatly reduce the constraints on microwell geometries and channel design, but also benefits from the intrinsic amenability of the pneumatic valve technique with device integration and automation. Thus we envision that the μAPPAD technology will provide a scalable and widely adaptable platform to promote the development of advanced lab-on-a-chip systems integrating microscale sample processing with dPCR for a broad scope of applications, such as single cell analysis of tumor heterogeneity and genetic profiling of circulating exosomes directly in clinical samples.


 Please wait while we load your content...
Please wait while we load your content...