MATE-Seq: microfluidic antigen-TCR engagement sequencing†
Abstract
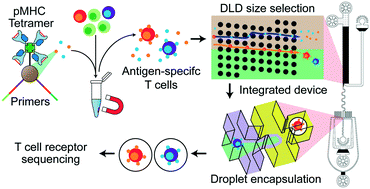
Adaptive immunity is based on peptide antigen recognition. Our ability to harness the immune system for therapeutic gain relies on the discovery of the T cell receptor (TCR) genes that selectively target antigens from infections, mutated proteins, and foreign agents. Here we present a method that selectively labels peptide antigen-specific CD8+ T cells using magnetic nanoparticles functionalized with peptide–MHC tetramers, isolates these specific cells within an integrated microfluidic device, and directly amplifies the TCR genes for sequencing. Critically, the identity of the peptide recognized by the TCR is preserved, providing the link between peptide and gene. The platform requires inputs on the order of just 100 000 CD8+ T cells, can be multiplexed for simultaneous analysis of multiple peptides, and performs sorting and isolation on chip. We demonstrate 1000-fold sensitivity enhancement of detecting antigen-specific TCRs relative to bulk analysis and simultaneous capture of two virus antigen-specific TCRs from a population of T cells.
- This article is part of the themed collection: Immunotherapy


 Please wait while we load your content...
Please wait while we load your content...