High throughput gene expression profiling of yeast colonies with microgel-culture Drop-seq†
Abstract
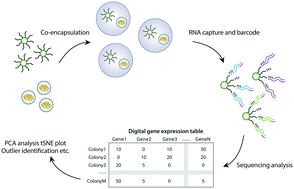
Yeast can be engineered into “living foundries” for non-natural chemical production by reprogramming them via a “design-build-test” cycle. While methods for “design” and “build” are relatively scalable and efficient, “test” remains a bottleneck, limiting the effectiveness of the procedure. Here we describe isogenic colony sequencing (ICO-seq), a massively-parallel strategy to assess the gene expression, and thus engineered pathway efficacy, of large numbers of genetically distinct yeast colonies. We use the approach to characterize opaque-white switching in 658 C. albicans colonies. By profiling the transcriptomes of 1642 engineered S. cerevisiae strains, we assess gene expression heterogeneity in a protein mutagenesis library. Our approach will accelerate synthetic biology by allowing facile and cost-effective transcriptional profiling of large numbers of genetically distinct yeast strains.
- This article is part of the themed collection: Droplet-based single-cell sequencing


 Please wait while we load your content...
Please wait while we load your content...