Droplet-based single cell RNAseq tools: a practical guide†
Abstract
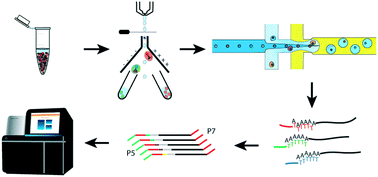
Droplet based scRNA-seq systems such as Drop-seq, inDrop and Chromium 10X have been the catalyst for the wide adoption of high-throughput scRNA-seq technologies in the research laboratory. In order to understand the capabilities of these systems to deeply interrogate biology; here we provide a practical guide through all the steps involved in a typical scRNA-seq experiment. Through comparing and contrasting these three main droplet based systems (and their derivatives), we provide an overview of all critical considerations in obtaining high quality and biologically relevant data. We also discuss the limitations of these systems and how they fit into the emerging field of Genomic Cytometry.
- This article is part of the themed collections: Lab on a Chip Recent Review Articles and Droplet-based single-cell sequencing


 Please wait while we load your content...
Please wait while we load your content...