Computational modeling of the catalytic mechanism of hydroxymethylbilane synthase†
Abstract
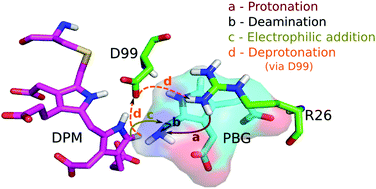
Hydroxymethylbilane synthase (HMBS), the third enzyme in the heme biosynthesis pathway, catalyzes the formation of 1-hydroxymethylbilane (HMB) by a stepwise polymerization of four molecules of porphobilinogen (PBG) using the dipyrromethane (DPM) cofactor. The mechanism by which HMBS polymerizes four units of PBG has not been elucidated to date. In vitro and in silico studies on HMBS have suggested certain residues with catalytic importance, but their specific role in the catalysis is unclear. To understand the catalytic mechanism of HMBS, quantum mechanical (QM) calculations were performed on model systems obtained from the active site of the human HMBS enzyme. The addition of one molecule of PBG to the DPM cofactor is carried out in four steps: (1) protonation of the substrate, PBG; (2) deamination of PBG; (3) electrophilic addition of the deaminated substrate to the terminal pyrrole ring of the enzyme-bound DPM cofactor and (4) deprotonation of the carbon atom at the α-position of the second ring of DPM. Based on the energy profiles from the QM calculations on cluster models, R26 is proposed to be the best suitable proton donor to the PBG moiety, which aids in the deamination of the substrate. During the electrophilic addition step, the intermediate formed is stabilized by the carboxylate side chain of the D99 residue. In the final deprotonation step, an extra proton from the second ring of DPM is transferred to R26 via the carboxylate side chain of D99, thus completing one cycle of the catalytic mechanism. The residues in the cluster model seem to play an important role in obtaining accurate energy barriers. All the stationary points along the reaction pathway have been characterized using QM calculations. The rate limiting step for the complete mechanism is found to be the deamination of the PBG moiety. The results of this study provide a detailed understanding of the catalytic mechanism and would help design future studies aimed at modulating the activity of HMBS.


 Please wait while we load your content...
Please wait while we load your content...