Nonnative contact effects in protein folding†
Abstract
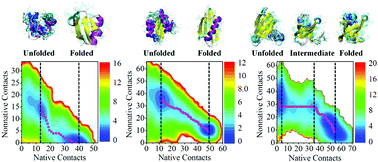
A comprehensive understanding of protein folding includes the knowledge of the formation of individual secondary structures, tertiary structure, and the effects of non-native contacts on these folding events. The measurement of these microscopic events has been posing challenges for experiment and molecular simulation. In this work, we performed enhanced sampling MD simulations for three proteins (NTL9, NuG2b, and CspA) and analyzed minimum free energy paths on multi-dimensional free energy landscapes to explore the underlying folding mechanisms. Consistencies can be seen between the present simulations and the existing experiments as well as other MD simulations. Quantitative analysis reveals the nucleation–condensation folding mechanism indicating the concurrent build-up of secondary and tertiary structures for the three proteins and gives the detailed formation sequence of individual native secondary structure elements. More importantly, nonnative contacts are generally observed among the proteins, creating a nonnative environment to affect the folding of individual secondary structure elements. A general tendency is that the secondary structure element(s) where the maximal nonnative contacts are observed have the largest formation free-energy barrier(s), corresponding to the rate-limiting step(s) of the folding for proteins that follow the nucleation–condensation mechanism. In summary, while native contacts determine the folding mechanism and pathway, non-native contacts play an important role in determining the protein folding thermodynamics by influencing the free energies of individual secondary structure element formation.


 Please wait while we load your content...
Please wait while we load your content...