A complex multiscale virtual particle model based elastic network model (CMVP-ENM) for the normal mode analysis of biomolecular complexes
Abstract
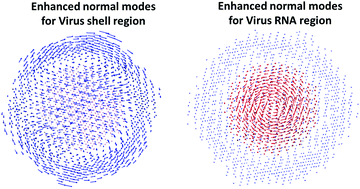
Understanding the molecular flexibility and dynamics is central to the analysis of biomolecular functions. In this work, complex multiscale virtual particle model based elastic network models (CMVP-ENMs) have been proposed for the normal mode analysis of biomolecular complexes or biomolecular assemblies. The term complex used in our CMVP-ENMs refers to the multi-material or multi-constituent. Different “materials” or constituents contribute differently to the general flexibility and dynamics of complex biomolecular structures. In our CMVP-ENMs, the key idea is to incorporate relative density or weight information of different components into the spring parameter of elastic network models. Two different models, including the CMVP based Gaussian network model (CMVP-GNM) and the CMVP based anisotropic network model (CMVP-ANM), have been proposed. With the consideration of complex component information, our CMVP-GNM, compared with the traditional GNM, can deliver a better accuracy in the B-factor prediction of protein–nucleic acid complexes. Moreover, our CMVP-ANM can be used to remove the “tip effect” by systematically suppressing the extremely-large vectors, in the highly flexible regions, of the normal modes generated by the ANM. In this way, our CMVP-ANM can be used to handle biomolecular structures with large hanging loops or extruding ends, which usually cause an irrationally-large-vector problem in ANM predictions. Finally, we explore the potential applications of our method by the cryo-EM data analysis. We find that by tuning the relative density ratio, we can systematically enhance or suppress the modes in different components, so that it can reveal the dynamics of the special regions that we are interested in.


 Please wait while we load your content...
Please wait while we load your content...