Molecular mechanism and binding free energy of doxorubicin intercalation in DNA†
Abstract
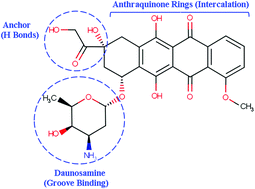
The intercalation process of binding doxorubicin (DOX) in DNA is studied by extensive MD simulations. Many molecular factors that control the binding affinity of DOX to DNA to form a stable complex are inspected and quantified by employing continuum solvation models for estimating the binding free energy. The modified MM-PB(GB)SA methodology provides a complete energetic profile of ΔGele, ΔGvDW, ΔGpolar, ΔGnon-polar, TΔStotal, ΔGdeform, ΔGcon, and ΔGion. To identify the sequence specificity of DOX, two different DNA sequences, d(CGATCG) or DNA1 and d(CGTACG) or DNA2, with one molecule (1 : 1 complex) or two molecule (2 : 1 complex) configurations of DOX were selected in this study. Our results show that the DNA deformation energy (ΔGdeform), the energy cost from translational and rotational entropic contributions (TΔStran+rot), the total electrostatic interactions (ΔGpolar-PB/GB + ΔGele) of incorporation, the intramolecular electrostatic interactions (ΔGele) and electrostatic polar solvation interactions (ΔGpolar-PB/GB) are all unfavorable to the binding of DOX to DNA. However, they are overcome by at least five favorable interactions: the van der Waals interactions (ΔGvDW), the non-polar solvation interaction (ΔGnon-polar), the vibrational entropic contribution (TΔSvib), and the standard concentration dependent free energies of DOX (ΔGcon) and the ionic solution (ΔGion). Specifically, the van der Waals interaction appears to be the major driving force to form a stable DOX–DNA complex. We also predict that DOX has stronger binding to DNA1 than DNA2. The DNA deformation penalty and entropy cost in the 2 : 1 complex are less than those in the 1 : 1 complex, thus they indicate that the 2 : 1 complex is more stable than the 1 : 1 complex. We have calculated the total binding free energy (BFE) (ΔGt-sim) using both MM-PBSA and MM-GBSA methods, which suggests a more stable DOX–DNA complex at lower ionic concentration. The calculated BFE from the modified MM-GBSA method for DOX–DNA1 and DOX–DNA2 in the 1 : 1 complex is −9.1 and −5.1 kcal mol−1 respectively. The same quantities from the modified MM-PBSA method are −12.74 and −8.35 kcal mol−1 respectively. The value of the total BFE ΔGt-sim in the 1 : 1 complex is in reasonable agreement with the experimental value of −7.7 ± 0.3 kcal mol−1.


 Please wait while we load your content...
Please wait while we load your content...