Enhancement of exon skipping activity by reduction in the secondary structure content of LNA-based splice-switching oligonucleotides†
Abstract
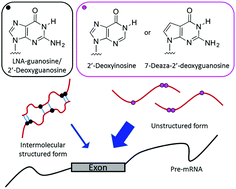
PAGE and UV melting analysis revealed that longer LNA-based splice-switching oligonucleotides (SSOs) formed secondary structures by themselves, reducing their effective concentration. To avoid such secondary structure formation, we introduced 7-deaza-2′-deoxyguanosine or 2′-deoxyinosine into the SSOs. These modified SSOs, with fewer secondary structures, showed higher exon skipping activities.


 Please wait while we load your content...
Please wait while we load your content...