Pre-lab proteolysis for dried serum spots – a paper-based sampling concept targeting low abundant biomarkers†
Abstract
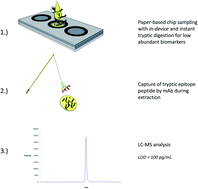
Paper-based sampling of biological matrices in combination with mass spectrometry has proven to be a promising technique for bottom-up analysis of proteins. However, these techniques have been challenging for determining low abundance proteins (pg mL−1 to low ng mL−1) due to low sampling volumes and adsorption of proteins to the sampling material. Here we are introducing a paper-based sampling concept in chip format with in-device proteolysis combined with selective proteotypic epitope peptide extraction. The sampling concept is demonstrated with the low abundant biomarker proGRP and showed promising performance from 500 pg mL−1 to 1000 ng mL−1 (R2 = 0.99) of proGRP spiked to human serum samples (all RSDs within 26%) in a five point concentration curve (n = 3). LOD and LOQ were determined to 150 pg mL−1 and 500 pg mL−1, respectively. The combination of paper-based sampling with in-device proteolysis and subsequent proteotypic epitope peptide immunocapture showed promising performance for other clinical applications.


 Please wait while we load your content...
Please wait while we load your content...