Profiling embryonic stem cell differentiation by MALDI TOF mass spectrometry: development of a reproducible and robust sample preparation workflow†
Abstract
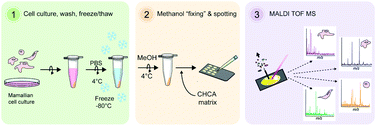
MALDI TOF mass spectrometry (MS) is widely used to characterise and biotype bacterial samples, but a complementary method for profiling of mammalian cells is still underdeveloped. Current approaches vary dramatically in their sample preparation methods and are not suitable for high-throughput studies. In this work, we present a universal workflow for mammalian cell MALDI TOF MS analysis and apply it to distinguish ground-state naïve and differentiating mouse embryonic stem cells (mESCs), which can be used as a model for drug discovery. We employed a systematic approach testing many parameters to evaluate how efficiently and reproducibly each method extracted unique mass features from four different human cell lines. These data enabled us to develop a unique mammalian cell MALDI TOF workflow involving a freeze–thaw cycle, methanol fixing and a CHCA matrix to generate spectra that robustly phenotype different cell lines and are highly reproducible in peak identification across replicate spectra. We applied our optimised workflow to distinguish naïve and differentiating populations using multivariate analysis and reproducibly identify unique features. We were also able to demonstrate the compatibility of our optimised method for current automated liquid handling technologies. Consequently, our MALDI TOF MS profiling method enables identification of unique features and robust phenotyping of mESC differentiation in under 1 hour from culture to analysis, which is significantly faster and cheaper when compared with conventional methods such as qPCR. This method has the potential to be automated and can in the future be applied to profile other cell types and expanded towards cellular MALDI TOF MS screening assays.


 Please wait while we load your content...
Please wait while we load your content...