Development of methionine methylation profiling and relative quantification in human breast cancer cells based on metabolic stable isotope labeling†
Abstract
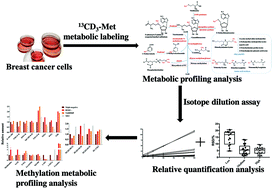
Methylation of components involved in one-carbon metabolism is extremely important in cancer; comprehensive studies on methylation are essential and may provide us with a better understanding of tumorigenesis, and lead to the discovery of potential biomarkers. Here, we present an improved methodology for methylated metabolite profiling and its relative quantification in breast cancer cell lines by isotope dilution mass spectrometry based on 13CD3-methionine metabolic labeling using ultra-high-performance liquid chromatography coupled with high-resolution tandem mass spectrometry (UPLC-HRMS/MS). First, all the methylated metabolites related to methionine were first screened and profiled by introducing 13CD3-methionine as the only medium into breast cancer cell growth cultures for both cellular polar metabolites and lipids. In total, we successfully found 20 labeled methylated metabolites and most of them were identified, some of which have not been reported before. We also developed a relative quantification method for all identified methylated metabolites based on isotope dilution mass spectrometry assays. Finally, the developed method was used for different breast cancer cells and mammary epithelial cells. Most methylated metabolites were disrupted in cancer cells. 1-Methyl-nicotinamide was decreased significantly, while trimethylglycine-glutamic acid-lysine and trimethyl-lysine were increased more than five times. This method offers a new insight into the methylation process, with several key pathways and important new metabolites being identified. Further investigation with biological assays should help to reveal the overall methylation metabolic network.


 Please wait while we load your content...
Please wait while we load your content...