Crossing constriction channel-based microfluidic cytometry capable of electrically phenotyping large populations of single cells
Abstract
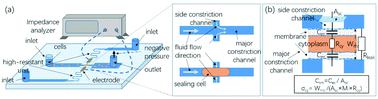
This paper presents a crossing constriction channel-based microfluidic system for high-throughput characterization of specific membrane capacitance (Csm) and cytoplasm conductivity (σcy) of single cells. In operations, cells in suspension were forced through the major constriction channel and instead of invading the side constriction channel, they effectively sealed the side constriction channel, which led to variations in impedance data. Based on an equivalent circuit model, these raw impedance data were translated into Csm and σcy. As a demonstration, the developed microfluidic system quantified Csm (3.01 ± 0.92 μF cm−2) and σcy (0.36 ± 0.08 S m−1) of 100 000 A549 cells, which could generate reliable results by properly controlling cell positions during their traveling in the crossing constriction channels. Furthermore, the developed microfluidic impedance cytometry was used to distinguish paired low- and high-metastatic carcinoma cell types of SACC-83 (ncell = ∼100 000) and SACC-LM cells (ncell = ∼100 000), distinguishing significant differences in both Csm (3.16 ± 0.90 vs. 2.79 ± 0.67 μF cm−2) and σcy (0.36 ± 0.06 vs.0.41 ± 0.08 S m−1). As high-throughput microfluidic impedance cytometry, this technique may add a new marker-free dimension to flow cytometry in single-cell analysis.
- This article is part of the themed collection: Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...