Single-molecule DNA visualization using AT-specific red and non-specific green DNA-binding fluorescent proteins†
Abstract
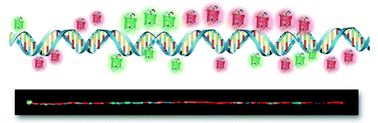
The recent advances in the single cell genome analysis are generating a considerable amount of novel insights into complex biological systems. However, there are still technical challenges because each cell has a single copy of DNA to be amplified in most single cell genome analytical methods. In this paper, we present a novel approach to directly visualize a genomic map on a large DNA molecule instantly stained with red and green DNA-binding fluorescent proteins without DNA amplification. For this visualization, we constructed a few types of fluorescent protein-fused DNA-binding proteins: H-NS (histone-like nucleoid-structuring protein), DNA-binding domain of BRCA1 (breast cancer 1), high mobility group-1 (HMG), and lysine tryptophan (KW) repeat motif. Because H-NS and HMG preferentially bind A/T-rich regions, we combined A/T specific binder (H-NS-mCherry and HMG-mCherry as red color) and a non-specific complementary DNA binder (BRCA1-eGFP and 2(KW)2-eGFP repeat as green color) to produce a sequence-specific two-color DNA physical map for efficient optical identification of single DNA molecules.
- This article is part of the themed collection: Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...