Assessing the performance of the MM/PBSA and MM/GBSA methods. 10. Impacts of enhanced sampling and variable dielectric model on protein–protein Interactions†
Abstract
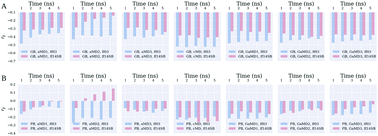
Enhanced sampling has been extensively used to capture the conformational transitions in protein folding, but it attracts much less attention in the studies of protein–protein recognition. In this study, we evaluated the impact of enhanced sampling methods and solute dielectric constants on the overall accuracy of the molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) and molecular mechanics/generalized Born surface area (MM/GBSA) approaches for the protein–protein binding free energy calculations. Here, two widely used enhanced sampling methods, including aMD and GaMD, and conventional molecular dynamics (cMD) simulations with two AMBER force fields (ff03 and ff14SB) were used to sample the conformations for 21 protein–protein complexes. The MM/PBSA and MM/GBSA calculation results illustrate that the standard MM/GBSA based on the cMD simulations yields the best Pearson correlation (rp = −0.523) between the predicted binding affinities and the experimental data, which is much higher than that given by MM/PBSA (rp = −0.212). Two enhanced sampling methods (aMD and GaMD) are indeed more efficient for conformational sampling, but they did not improve the binding affinity predictions for protein–protein systems, suggesting that the aMD or GaMD sampling (at least in short timescale simulations) may not be a good choice for the MM/PBSA and MM/GBSA predictions of protein–protein complexes. The solute dielectric constant of 1.0 is recommended to MM/GBSA, but a higher solute dielectric constant is recommended to MM/PBSA, especially for the systems with higher polarity on the protein–protein binding interfaces. Then, a preliminary assessment of the MM/GBSA calculations based on a variable dielectric generalized Born (VDGB) model was conducted. The results highlight the potential power of VDGB in the free energy predictions for protein–protein systems, but more thorough studies should be done in the future.
- This article is part of the themed collection: 2019 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...