Analysis of long dsRNA produced in vitro and in vivo using atomic force microscopy in conjunction with ion-pair reverse-phase HPLC†
Abstract
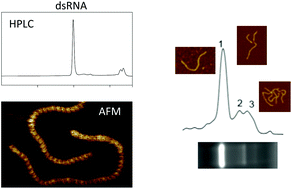
Long double-stranded (ds) RNA is emerging as a novel alternative to chemical and genetically-modified insect and fungal management strategies. The ability to produce large quantities of dsRNA in either bacterial systems, by in vitro transcription, in cell-free systems or in planta for RNA interference applications has generated significant demand for the development and application of analytical tools for analysis of dsRNA. We have utilised atomic force microscopy (AFM) in conjunction with ion-pair reverse-phase high performance liquid chromatography (IP-RP-HPLC) to provide novel insight into dsRNA for RNAi applications. The AFM analysis enabled direct structural characterisation of the A-form duplex dsRNA and accurate determination of the dsRNA duplex length. Moreover, further analysis under non-denaturing conditions revealed the presence of heterogeneous dsRNA species. IP-RP-HPLC fractionation and AFM analysis revealed that these alternative RNA species do not arise from different lengths of individual dsRNA molecules in the product, but represent misannealed RNA species that present as larger assemblies or multimeric forms of the RNA. These results for the first time provide direct structural insight into dsRNA produced both in vivo in bacterial systems and in vitro, highlighting the structural heterogeneity of RNA produced. These results are the first example of detailed characterisation of the different forms of dsRNA from two production systems and establish atomic force microscopy as an important tool for the characterisation of long dsRNA.


 Please wait while we load your content...
Please wait while we load your content...