Single-stranded DNA oligomer brush structure is dominated by intramolecular interactions mediated by the ion environment
Abstract
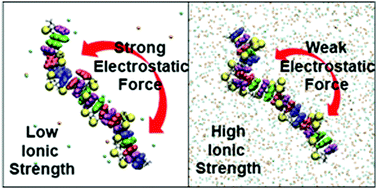
Single-stranded DNA (ssDNA) brushes, in which ssDNA oligomers are tethered to surfaces in dense monolayers, are being investigated for potential biosensing applications. The structure of the brush can affect the selectivity and the hybridization efficiency of the device. The structure is commonly thought to result from the balance of intramolecular interactions, intermolecular interactions within the monolayer, and molecule–surface interactions. Here, we test the hypothesis that ssDNA oligomer brush structure is dominated by intramolecular interactions. We use AFM to measure the height of an ssDNA brush and molecular dynamics to simulate the end-to-end distance, both as a function of ionic strength of the surrounding solution. The brush height and the molecule end-to-end distance match quantitatively, providing evidence that the brush structure is dominated by intramolecular interactions (mediated by ions). The physical basis of the intramolecular interactions is elucidated by the simulations.


 Please wait while we load your content...
Please wait while we load your content...