Design and virtual screening of novel fluoroquinolone analogs as effective mutant DNA GyrA inhibitors against urinary tract infection-causing fluoroquinolone resistant Escherichia coli†
Abstract
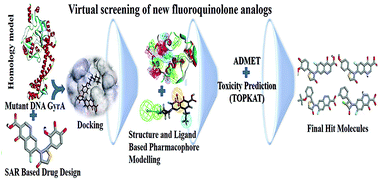
Fluoroquinolones (FQs) belong to the class of quinolone drugs that are used to treat Urinary tract infections (UTIs) through inhibition of E. coli DNA gyrase. Resistance to FQs poses a serious problem in the treatment against resistant strains of E. coli which are associated with Ser83 to Leu and Asp87 to Asn mutations at the quinolone resistance determining region (QRDR) of the GyrA subunit of DNA gyrase. Mutant DNA GyrA (mtDNA GyrA) is deemed to be a significant target for the development of novel FQ drugs. Due to resistance to FQ drugs, discovery or development of novel FQs is crucial to inhibit the mtDNA GyrA. Hence, the present study attempts to design and develop novel FQs that are efficient against resistant E. coli strains. A three-dimensional structure of the mtDNA GyrA protein was developed by homology modeling, following which 204 novel FQ analogs were designed using target based SAR. The designed ligands were then screened using molecular docking studies, through which the pattern of interaction between the ligands and the target protein was studied. As expected, the results of the docking study revealed that the molecules FQ-147, FQ-151 and FQ-37 formed hydrogen bonding and Van der Waals interactions with Leu83 and Asn87 (mutated residues), respectively. Further, the wild-type (WT), mtDNA GyrA and docking complex were studied by molecular dynamics (MD) simulations. Subsequently, all the screened compounds were subjected to a structure and ligand based pharmacophore study followed by ADMET and toxicity (TOPKAT) prediction. Finally, eighteen hit FQ analogs which showed good results for the following properties, viz., best binding score, estimated activity (MIC value) and calculated drug-like properties, and least toxicity, were shortlisted and identified as potential leads to treat UTI caused by FQ resistant E. coli. Apart from development of novel drug candidates for inhibition of mtDNA GyrA, the present study also contributes towards a superior comprehension of the interaction pattern of ligands in the target protein. To a more extensive degree, the present work will be useful for the rational design of novel and potent drugs for UTIs.
- This article is part of the themed collection: Editors' Collection: Fluorine chemistry in medicinal chemistry and chemical biology


 Please wait while we load your content...
Please wait while we load your content...