Efficiency and fidelity of T3 DNA ligase in ligase-catalysed oligonucleotide polymerisations†
Abstract
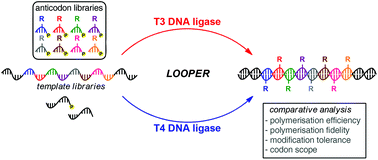
Ligase-catalyzed oligonucleotide polymerisations (LOOPER) can readily generate libraries of diversely-modified nucleic acid polymers, which can be subjected to iterative rounds of in vitro selection to evolve functional activity. While there exist several different DNA ligases, T4 DNA ligase has most often been used for the process. Recently, T3 DNA ligase was shown to be effective in LOOPER; however, little is known about the fidelity and efficiency of this enzyme in LOOPER. In this paper we evaluate the efficiency of T3 DNA ligase and T4 DNA ligase for various codon lengths and compositions within the context of polymerisation fidelity and yield. We find that T3 DNA ligase exhibits high efficiency and fidelity with short codon lengths, but struggles with longer and more complex codon libraries, while T4 DNA ligase exhibits the opposite trend. Interestingly, T3 DNA ligase is unable to accommodate modifications at the 8-position of adenosine when integrated into short codons, which will create challenges in expanding the available codon set for the process. The limitations and strengths of the two ligases are further discussed within the context of LOOPER.
- This article is part of the themed collections: Chemical Biology in OBC and New Talent


 Please wait while we load your content...
Please wait while we load your content...