Target-initiated synthesis of fluorescent copper nanoparticles for the sensitive and label-free detection of bleomycin†
Abstract
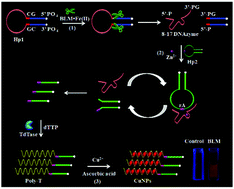
Fluorescent copper nanoparticles (CuNPs) have received great attention due to their distinct characteristics of facile synthesis, tunable fluorescence emission, high photostability, and biological compatibility, and they have been widely used for chemical and biological analyses. Bleomycins (BLMs) are widely used antitumor agents for the clinical treatment of various cancers. Here, we develop a sensitive and label-free fluorescence method for the quantitative detection of BLM on the basis of BLM-initiated enzymatic polymerization-mediated synthesis of fluorescent CuNPs. We design two hairpin DNAs: one (Hp1) for the recognition of BLM and the other (Hp2) for signal amplification. In the presence of BLM, it may recognize and cleave the 5′-GC-3′ site of the Hp1 stem, releasing the 8–17 DNAzyme fragment. The resultant 8–17 DNAzyme fragments may bind with the loop of Hp2 to form a partial double-stranded DNA (dsDNA) duplex, initiating the cyclic cleavage of Hp2 in the presence of Zn2+-dependent DNAzymes and generating numerous new DNA fragments with the free 3′-OH terminal, which can induce the formation of a poly(thymine) (poly-T) sequence with the assistance of terminal deoxynucleotidyl transferase (TdTase). Subsequently, the ploy-T sequence may function as the template for the synthesis of CuNPs with strong fluorescence emission. This method shows good selectivity and high sensitivity with a detection limit as low as 8.1 × 10−16 M, and it exhibits good performance in serum samples. Moreover, this method has distinct advantages of simplicity and low cost, holding great potential in clinical diagnosis and biomedical research.


 Please wait while we load your content...
Please wait while we load your content...