Effective lock-in strategy for proteomic analysis of corona complexes bound to amino-free ligands of gold nanoparticles†
Abstract
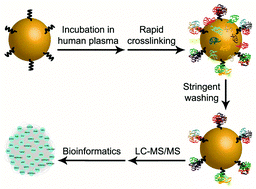
For specific applications, gold nanoparticles (GNPs) are commonly functionalized with various biological ligands, including amino-free ligands such as amino acids, peptides, proteins, and nucleic acids. Upon entering a biological fluid, the protein corona that forms around GNPs can conceal the targeting ligands and sterically hinder the functional properties. The protein corona is routinely prepared by standard centrifugation or sucrose cushion centrifugation. However, such methodologies are not applicable to the exclusive analysis of a ligand-binding protein corona. In this study, we first proposed a lock-in strategy based on a combination of rapid crosslinking and stringent washing. Cysteine was used as a model of amino-free ligands and attached to GNPs. After corona formation in the human plasma, GNP cysteine and corona proteins were quickly fixed by 5 s of crosslinking with 7.5% formaldehyde. After stringent washing using SDS buffer with sonication, the cysteine-bound proteins were effectively separated from unbound proteins. Qualitative and quantitative analyses using a mass spectrometry-based proteomics approach indicated that the protein composition of the cysteine-binding corona from the new method was significantly different from the composition of the whole corona from the two conventional methods. Furthermore, network and formaldehyde-linked site analyses of cysteine-binding proteins provided useful information toward a better knowledge of the behavior of protein–ligand and protein–protein interactions. Collectively, our new strategy has the capability to particularly characterize the protein composition of a cysteine-binding corona. The presented methodology in principal provides a generic way to analyze a nanoparticle corona bound to amino-free ligands and has the potential to decipher corona-masked ligand functions.


 Please wait while we load your content...
Please wait while we load your content...