Modeling of canonical and C2′-O-thiophenylmethyl modified hexamers of RNA. Insights into the nature of structural changes and thermal stability†
Abstract
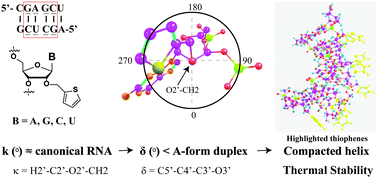
Modifying the ribose ring or the nucleobase within oligonucleotides of RNA is a strategy that has regained attention due to its potential to provide chemical stability, increase affinity towards a target, or introduce groups with bioorthogonal reactivity, among other applications. However introducing groups via synthetic routes can be a daunting process and may result in unexpected physical properties and structural variations. In addition, characterizing the effects of a modification within an oligonucleotide can be time consuming and expensive, e.g., NMR, crystallography. Thus the development of models that are accurate, reasonably priced and carried out over short periods of time is essential to the development of this field. Herein we report on the development and validation of computational models that explain the nature of the structural and stability changes induced by the presence of a C2′-O-methylthiophene group at various positions within duplexes conformed by two 6-mers of modified RNA as well as random coils containing the same modification(s). Through analysis of the obtained models it was concluded that the presence of the methylthiophene moiety on two strands of RNA induces a conformational change around the torsion angle (κ°) that leads to lower values in the pseudorotational angle (δ°), thus resulting in an A-form α-helix with shorter distances between phosphates than those in canonical RNA. These results provide a reasonable explanation for the experimental observations, which display the thermal stabilization of the duplexes, and are guiding efforts from our groups to control the structure and function of RNA, i.e., via functionalization of the RNA structure for attaining thermal stability while retaining the structural properties of an A-form duplex.


 Please wait while we load your content...
Please wait while we load your content...
