Handling of nutrient copper in the bacterial envelope
Abstract
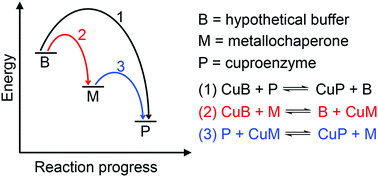
In bacteria, copper (Cu) is often recognised for its potential toxicity and its antibacterial activity is now considered a key component of the mammalian innate immune system. Cu ions bound in weak sites can catalyse harmful redox reactions while Cu ions in strong but adventitious sites can disrupt protein or enzyme function. For these reasons, the outward transport of Cu from bacteria has received significant attention. Yet, Cu is also a bacterial nutrient, required as a cofactor by enzymes that catalyse electron transfer processes, for instance in aerobic and anaerobic respiration. To date, the inward flow of this metal ion as a nutrient and its insertion into target cuproenzymes remain poorly defined. Here we revisit the available evidence related to bacterial nutrient Cu trafficking and identify gaps in knowledge. Particularly intriguing is the evidence that bacterial cuproenzymes do not always require auxiliary metallochaperones to insert nutrient Cu into their active sites. This review outlines our effort to consolidate the available experimental data using an established energy-driven model for metalation.
- This article is part of the themed collections: Metallomics Recent HOT articles, Recent Review Articles and Metallomics Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...