Statistical analysis of multi-dimensional, temporal gene expression of stem cells to elucidate colony size-dependent neural differentiation†
Abstract
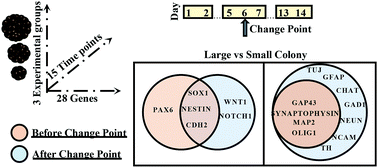
High throughput gene expression analysis using qPCR is commonly used to identify molecular markers of complex cellular processes. However, statistical analysis of multi-dimensional, temporal gene expression data is complicated by limited biological replicates and large number of measurements. Moreover, many available statistical tools for analysis of time series data assume that the data sequence is static and does not evolve over time. With this assumption, the parameters used to model the time series are fixed and thus, can be estimated by pooling data together. However, in many cases, dynamic processes of biological systems involve abrupt changes at unknown time points, making the assumption of stationary time series break down. We addressed this problem using a combination of statistical methods including hierarchical clustering, change point detection, and multiple testing. We applied this multi-step method to multi-dimensional, temporal gene expression data that resulted from our study of colony size-dependent neural cell differentiation of stem cells. The gene expression data were time series as the observations were recorded sequentially over time. Hierarchical clustering segregated the genes into three distinct clusters based on their temporal expression profiles; change point detection identified specific time points at which the entire dataset was divided into several homogenous subsets to allow a separate analysis of each subset; and multiple testing procedure identified the differentially expressed genes in each cluster within each subset of data. We established that our multi-step approach pinpoints specific sets of genes that underlie colony size-mediated neural differentiation of stem cells and demonstrated its advantages over conventional parametric and non-parametric tests that do not take into account temporal dynamics of the data. Importantly, our proposed approach is broadly applicable to any multivariate data sets of limited sample size from high throughput and high content screening such as in drug and biomarker discovery studies.


 Please wait while we load your content...
Please wait while we load your content...