A toolbox for discrete modelling of cell signalling dynamics†
Abstract
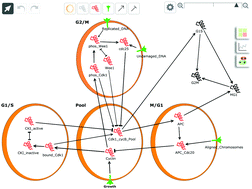
In an age where the volume of data regarding biological systems exceeds our ability to analyse it, many researchers are looking towards systems biology and computational modelling to help unravel the complexities of gene and protein regulatory networks. In particular, the use of discrete modelling allows generation of signalling networks in the absence of full quantitative descriptions of systems, which are necessary for ordinary differential equation (ODE) models. In order to make such techniques more accessible to mainstream researchers, tools such as the BioModelAnalyzer (BMA) have been developed to provide a user-friendly graphical interface for discrete modelling of biological systems. Here we use the BMA to build a library of discrete target functions of known canonical molecular interactions, translated from ordinary differential equations (ODEs). We then show that these BMA target functions can be used to reconstruct complex networks, which can correctly predict many known genetic perturbations. This new library supports the accessibility ethos behind the creation of BMA, providing a toolbox for the construction of complex cell signalling models without the need for extensive experience in computer programming or mathematical modelling, and allows for construction and simulation of complex biological systems with only small amounts of quantitative data.


 Please wait while we load your content...
Please wait while we load your content...