Consequences of Mg2+ binding on the geometry and stability of RNA base pairs†
Abstract
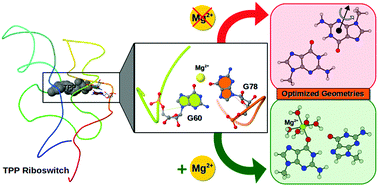
Metal ions are crucial for folding and function of noncoding RNAs. The fact that RNAs have very specific metal ion binding motifs further implies that contribution of metal ions (like Mg2+) in RNA's folding is not limited to simple compensation of electrostatic repulsions. Rather, their binding to RNA is driven by very specific contextual requirements. Elucidation of such factors is necessary for a comprehensive understanding of the sequence–structure–function paradigm in RNA. In this work, we have studied the consequences of Mg2+ binding on the geometry and stability of different noncanonical base pairs that shape up the complex structural landscape of RNA. Our results show that majority of the Mg2+ bound nucleobases are also part of a base pair. Interestingly, such base pairs belong only to a specific set of base pairing geometries. Out of them, we are able to identify 14 unique cases for which the native base pairing geometries are unstable under gas phase geometry optimization carried out in the absence of Mg2+ binding. Our density functional theory based calculations, performed using dispersion corrected M05-2X functional, suggest that, depending on its mode of binding, Mg2+ can stabilize and even fine tune a number of such base pairing geometries. These findings not only provide insights into how metal ions modulate the structure and dynamics of RNA molecules, they also provide a basis for improving the RNA structure prediction algorithms.


 Please wait while we load your content...
Please wait while we load your content...