Enhanced basepair dynamics pre-disposes protein-assisted flips of key bases in DNA strand separation during transcription initiation†
Abstract
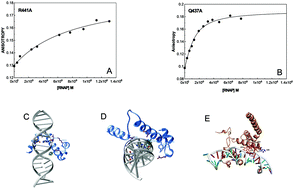
Localized separation of strands of duplex DNA is a necessary step in many DNA-dependent processes, including transcription and replication. Little is known about how these strand separations occur. The strand-separated E.coli RNA polymerase–promoter open-complex structure showed four bases of the non-template strand, the master base −11A, −7, −6 and +2, in a flipped state and inserted into protein pockets. To explore whether any property of these bases in the duplex state pre-disposes them to flipping, NMR studies were performed on a wild-type promoter in the duplex state. Measurement of relaxation times indicates that a limited number of base pairs, including the flipped ones, have faster opening rates than the rest. Molecular dynamics studies also show an inherently high dynamic character of the −11A:T base pair in the wild-type strand-paired state. In order to explore the role of the RNA polymerase in the flipping process, we have used 2-aminopurine as a fluorescent probe. Slower kinetics of the increase of 2-aminopurine fluorescence was observed with RNA polymerases containing several mutant σ70s. This may be interpreted as the protein playing an important role in enhancing the flipping rate. These results suggest that flipping of −11A, and perhaps other flipped bases observed in the open-complex, is facilitated by its inherent proclivity to open-up with further assistance from the protein, thus leading to a strand-open state. Other DNA-based processes that require strand-separation may use similar pathways for strand separation. We conclude that not only basepair stability, but also dynamics may play an important role in the strand-separation.


 Please wait while we load your content...
Please wait while we load your content...