Nucleobases-decorated boron nitride nanoribbons for electrochemical biosensing: a dispersion-corrected DFT study†
Abstract
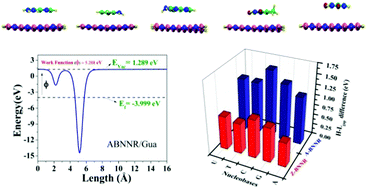
Understanding the interactions between biomolecules and boron nitride nanostructures is key for their use in nanobiotechnology and medical engineering. In this study, we investigated the adsorption of nucleobases adenine (A), guanine (G), cytosine (C), thymine (T) and uracil (U) over armchair and zigzag boron nitride nanoribbons (BNNR) using density functional theory to define the applicability of BNNR for the sensing of nucleobases and DNA sequencing. To appropriately account for dispersion, the van der Waals forces (DFT-D2)-type method developed by Grimme was also included in the calculations. The calculated adsorption energy suggests the following order of adsorption for A-BNNR and Z-BNNR with the nucleobases: G > T > A > U > C and G > C > A > T > U, respectively. The origin of the binding of the different nucleobases with BNNR was analysed and π–π stacking was found to be responsible. In addition, the electronic properties, density of states and work function significantly vary after adsorption. These analyses indicate different binding natures for different nucleobases and BNNRs. Thus, this study demonstrates that BNNR can be applied as biosensors for the detection of nucleobases, which are constituents of DNA and RNA. Furthermore, analysis of electronic properties and adsorption energies will play a key role in targeted drug delivery, enzyme activities and genome sequencing. Our results indicate that BNNRs have better adsorption capacity than graphene and boron nitride nanotubes.


 Please wait while we load your content...
Please wait while we load your content...