Influence of the DNA sequence/length and pH on deaminase activity, as well as the roles of the amino acid residues around the catalytic center of APOBEC3F†
Abstract
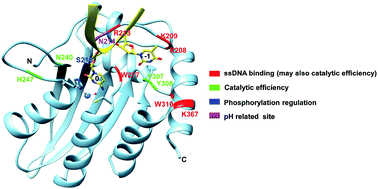
APOBEC3F (A3F), an apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) family protein, catalyzes cytosine-to-uracil conversion in single-stranded (ss) DNA. A3F acts as an inhibitor of retrovirus replication and exhibits antiviral activity against viral infectivity factor (Vif)-deficient human immunodeficiency virus 1 (HIV-1). Previous studies have mostly been focused on the interaction between A3F and Vif, and the studies on A3F's deamination properties are limited. Here, we report comprehensive characterization of the deaminase activity and ssDNA binding of the C-terminal domain (CTD) of A3F. It was shown that the deaminase activity of A3F-CTD is affected by the nucleic acid residues adjacent to the target sequence, TC, and that TTCA/G are the most preferred sequences. A3F-CTD deaminates the target sequence in longer ssDNAs most efficiently. Mutation analysis identified the amino acid residues that are responsible for the deaminase activity and ssDNA binding in the loops surrounding the catalytic center. The functions of these residues were rationally interpreted on the basis of the co-crystal structure of A3A-ssDNA and the known roles of the equivalent amino acid residues found in other A3s. Furthermore, we demonstrated that the deaminase activity of A3F-CTD could be regulated through phosphorylation of a putative site, S216. Finally, A3F-CTD was found to be active in a wide pH range (5.5 to 9.5) with similar activity. Interestingly, the A3F-CTD N214H mutant exhibited a dramatic increase in activity at pH 5.5.
- This article is part of the themed collection: Complex molecular systems: supramolecules, biomolecules and interfaces


 Please wait while we load your content...
Please wait while we load your content...