Investigation of compacted DNA structures induced by Na+ and K+ monovalent cations using biological nanopores†
Abstract
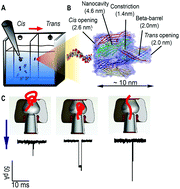
In aqueous solutions, an elongated, negatively charged DNA chain can quickly change its conformation into a compacted globule in the presence of positively charged molecules, or cations. This well-known process, called DNA compaction, is a method with great potential for gene therapy and delivery. Experimental conditions to induce these compacted DNA structures are often limited to the use of common compacting agents, such as cationic surfactants, polymers, and multivalent cations. In this study, we show that in highly concentrated buffers of 1 M monovalent cation solutions at pH 7.2 and 10, biological nanopores allow real-time sensing of individual compacted structures induced by K+ and Na+, the most abundant monovalent cations in human bodies. Herein, we studied the ratio between compacted and linear structures for 15-mer single-stranded DNA molecules containing only cytosine nucleotides, optimizing the probability of linear DNA chains being compacted. Since the binding affinity of each nucleotide to cation is different, the ability of the DNA strand to fold into a compacted structure greatly depends on the type of cations and nucleotides present. Our experimental results compare favorably with findings from previous molecular dynamics simulations for the DNA compacting potential of K+ and Na+ monovalent cations. We estimate that the majority of single-stranded DNA molecules in our experiment are compacted. From the current traces of nanopores, the ratio of compacted DNA to linear DNA molecules is approximately 30 : 1 and 15 : 1, at a pH of 7.2 and 10, respectively. Our comparative studies reveal that Na+ monovalent cations have a greater potential of compacting the 15C-ssDNA than K+ cations.
- This article is part of the themed collection: Analyst Recent HOT articles


 Please wait while we load your content...
Please wait while we load your content...