Kinetic resolution of (RS)-1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol: a metoprolol intermediate and its validation through homology model of Pseudomonas fluorescens lipase†
Abstract
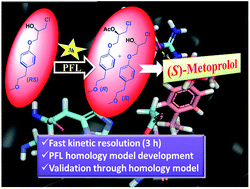
In the present study Pseudomonas fluorescens lipase (PFL) was screened as a time efficient biocatalyst for the kinetic resolution of a racemic intermediate [(RS)-1-chloro-3-(4-(2-methoxyethyl)phenoxy)propan-2-ol] of metoprolol, an important selective β1-blocker drug. PFL selectively acylated the R-form of this racemic intermediate in a short duration of 3 h. Different reaction parameters were optimized to achieve maximum enantioselectivity. It was found that at 30 °C, enzyme activity of 400 units and substrate concentration of 10 mM gave a high enantioselectivity and conversion in an optimum time of 3 hours (C = 50.5%, eep = 97.2%, ees = 95.4%, E = 182). To validate these experimental results, the 3D structure of PFL was built using homology modelling. Validation of the model through Ramachandran plot (92.7% in favored region), Errat plot (overall quality factor, 79.27%), Verify-3D score (86.19) and ProSA_Z score (−6.24) depicted the overall good quality of the model. Molecular docking of the R- and S-enantiomers of the intermediate, which was performed on this model, demonstrated a strong H-bond interaction (1.6 Å) between the hydroxyl group of the R-enantiomer and Arg54, a key binding residue of the catalytic site of PFL, while no significant interaction with the S-enantiomer was observed. Thus, the outcome of this docking study was in agreement with the experimental data, clarifying that PFL preferentially catalysed the transesterification of the R-enantiomer into the corresponding ester, leaving the S-enantiomer intact.


 Please wait while we load your content...
Please wait while we load your content...