Substoichiometric ribose methylations in spliceosomal snRNAs†
Abstract
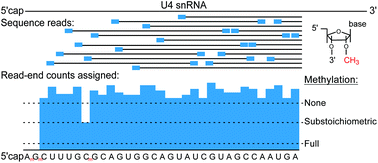
Sequencing-based profiling of ribose methylations is a new approach that allows for experiments addressing dynamic changes on a large scale. Here, we apply such a method to spliceosomal snRNAs present in human whole cell RNA. Analysis of solid tissue samples confirmed all previously known sites and demonstrated close to full methylation at almost all sites. Methylation changes were revealed in biological experimental settings, using T cell activation as an example, and in the T cell leukemia model, Jurkat cells. Such changes could impact the dynamics of snRNA interactions during the spliceosome cycle and affect mRNA splicing efficiency and splicing patterns.
- This article is part of the themed collection: Nucleic Acid Modifications


 Please wait while we load your content...
Please wait while we load your content...