The competing effects of core rigidity and linker flexibility in the nanoassembly of trivalent small molecule-DNA hybrids (SMDH3s)–a synergistic experimental-modeling study†
Abstract
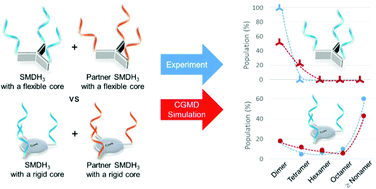
The nanoassembly behavior of trivalent small molecule-DNA hybrids (SMDH3s) was investigated as a function of core geometry and supramolecular flexibility through a synergistic experimental-modeling study. While complementary SMDH3s possessing a highly flexible tetrahedral trivalent core primarily assemble into nanoscale caged dimers, the nanoassemblies of SMDH3 comonomers with rigid pyramidal and trigonal cores yield fewer caged dimers and more large-oligomer networks. Specifically, the rigid pyramidal SMDH3 comonomers tend to form smaller nanosized aggregates (dimers, tetramers, and hexamers) upon assembly, attributable to the small (<109°) branch-core-branch angle of the pyramidal core. In contrast, the more-rigid trigonal planar SMDH3 comonomers have a larger (∼120°) branch-core-branch angle, which spaces their DNA arms farther apart, facilitating the formation of larger nanoassemblies (≥nonamers). The population distributions of these nanoassemblies were successfully captured by coarse-grained molecular dynamics (CGMD) simulations over a broad range of DNA concentrations. CGMD simulations can also forecast the effect of incorporating Tn spacer units between the hydridizing DNA arms and the rigid organic cores to increase the overall flexibility of the SMDH3 comonomers. Such “decoupling” of the DNA arms from the organic core was found to result in preferential formation of nanoscale dimers up to an optimal spacer length, beyond which network formation takes over due to entropic factors. This excellent agreement between the simulation and experimental results confirms the versatility of the CGMD model as a useful and reliable tool for elucidating the nanoassembly of SMDH-based building blocks.


 Please wait while we load your content...
Please wait while we load your content...