A systematic analysis of atomic protein–ligand interactions in the PDB†
Abstract
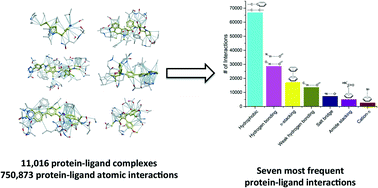
As the protein databank (PDB) recently passed the cap of 123 456 structures, it stands more than ever as an important resource not only to analyze structural features of specific biological systems, but also to study the prevalence of structural patterns observed in a large body of unrelated structures, that may reflect rules governing protein folding or molecular recognition. Here, we compiled a list of 11 016 unique structures of small-molecule ligands bound to proteins – 6444 of which have experimental binding affinity – representing 750 873 protein–ligand atomic interactions, and analyzed the frequency, geometry and impact of each interaction type. We find that hydrophobic interactions are generally enriched in high-efficiency ligands, but polar interactions are over-represented in fragment inhibitors. While most observations extracted from the PDB will be familiar to seasoned medicinal chemists, less expected findings, such as the high number of C–H⋯O hydrogen bonds or the relatively frequent amide–π stacking between the backbone amide of proteins and aromatic rings of ligands, uncover underused ligand design strategies.


 Please wait while we load your content...
Please wait while we load your content...