Identification of perturbed signaling pathways from gene expression data using information divergence†
Abstract
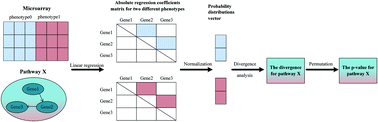
Abnormal regulation of signaling pathways is the key causative factor in several diseases. Although many methods have been proposed to identify significantly differential pathways between two conditions via microarray gene expression datasets, most of them concentrate on differences in the pathway components—either the differential expression or the correlation of genes in a given pathway. However, as biological functional units, signaling pathways may have diverse activity patterns across different biological contexts. In order to detect overall changes in pathways, we propose an analysis model called SPAID (Signaling Pathway Analysis based on Information Divergence). SPAID is based on the concept of information divergence, which can be used to compare two conditions by computing the differential probability distribution of the regulation capacity. We compared SPAID with several classical algorithms using different datasets, and the results indicate that SPAID produces higher repeatability, has better performance and universality, and extracts more comprehensive information regarding the underlying biological processes. In conclusion, by introducing the idea of information divergence, our study measures differences in pathways from an overall perspective and will provide a complementary analysis framework for pathway analysis.


 Please wait while we load your content...
Please wait while we load your content...