GeneSPIDER – gene regulatory network inference benchmarking with controlled network and data properties†
Abstract
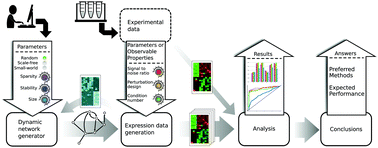
A key question in network inference, that has not been properly answered, is what accuracy can be expected for a given biological dataset and inference method. We present GeneSPIDER – a Matlab package for tuning, running, and evaluating inference algorithms that allows independent control of network and data properties to enable data-driven benchmarking. GeneSPIDER is uniquely suited to address this question by first extracting salient properties from the experimental data and then generating simulated networks and data that closely match these properties. It enables data-driven algorithm selection, estimation of inference accuracy from biological data, and a more multifaceted benchmarking. Included are generic pipelines for the design of perturbation experiments, bootstrapping, analysis of linear dependence, sample selection, scaling of SNR, and performance evaluation. With GeneSPIDER we aim to move the goal of network inference benchmarks from simple performance measurement to a deeper understanding of how the accuracy of an algorithm is determined by different combinations of network and data properties.


 Please wait while we load your content...
Please wait while we load your content...