Removal of bacterial cells, antibiotic resistance genes and integrase genes by on-site hospital wastewater treatment plants: surveillance of treated hospital effluent quality†
Abstract
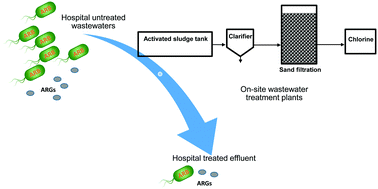
This study aims to evaluate the removal efficiency of microbial contaminants, including total cell counts, antibiotic-resistant bacteria (ARB), antibiotic resistance genes (ARGs, e.g. tetO, tetZ, sul1 and sul2) and integrase genes (e.g. intl1 and intl2), by wastewater treatment plants (WWTPs) operated on-site of two hospitals (i.e., SH WWTP and IH WWTP). Both SH and IH WWTPs utilize the conventional activated sludge process but differences in the removal efficiencies were observed. Over the 2 week sampling period, IH WWTP outperformed SH WWTP, and achieved an approximate 0.388 to 2.49-log log removal values (LRVs) for total cell counts compared to the 0.010 to 0.162-log removal in SH WWTP. Although ARB were present in the hospital influent, the treatment process of both hospitals effectively removed ARB from most of the effluent samples. In instances where ARB were recovered in the effluent, none of the viable isolates were identified to be opportunistic pathogenic species based on 16S rRNA gene sequencing. However, sul1 and intl1 genes remained detectable at up to 105 copies per mL or 8 × 10−1 copies per 16S rRNA gene in the treated effluent, with an LRV of less than 1.2. When the treated effluent is discharged from hospital WWTPs into the public sewer for further treatment as per requirement in many countries, the detected amount of ARGs and integrase genes in the hospital effluent can become a potential source of horizontal gene dissemination in the municipal WWTP. Proper on-site wastewater treatment and surveillance of the effluent quality for emerging contaminants are therefore highly recommended.


 Please wait while we load your content...
Please wait while we load your content...