A single-round selection of selective DNA aptamers for mammalian cells by polymer-enhanced capillary transient isotachophoresis†
Abstract
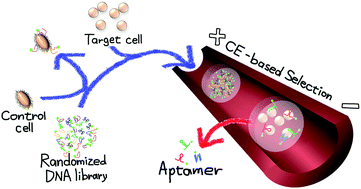
A single-round DNA aptamer selection for mammalian cells was successfully achieved for the first time using a capillary electrophoresis (CE)-based methodology called polymer-enhanced capillary transient isotachophoresis (PectI). The PectI separation yielded a single peak for the human lung cancer cell line (PC-9) complexed with DNA aptamer candidates, which was effectively separated from a free randomized DNA library peak, ensuring no contamination from free DNA in the PC-9–DNA aptamer complex fraction. The DNA aptamer candidates obtained after a single-round selection employing counter selection with HL-60 were proven to bind selectively and form kinetically stable complexes with PC-9 cells. Interestingly, most aptamer candidates showed high binding ability (Kd = 70–350 nM) with different extents of binding on the cell surface. These facts proved that a single-round selection for mammalian cells by PectI is feasible to obtain various types of aptamer candidates, which have high-affinity even for non-overexpressed but unique targets on the cell surface in addition to overexpressed targets.


 Please wait while we load your content...
Please wait while we load your content...