High-throughput screening of microchip-synthesized genes in programmable double-emulsion droplets†
Abstract
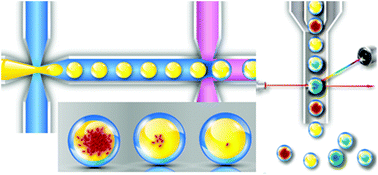
The rapid advances in synthetic biology and biotechnology are increasingly demanding high-throughput screening technology, such as screening of the functionalities of synthetic genes for optimization of protein expression. Compartmentalization of single cells in water-in-oil (W/O) emulsion droplets allows screening of a vast number of individualized assays, and recent advances in automated microfluidic devices further help realize the potential of droplet technology for high-throughput screening. However these single-emulsion droplets are incompatible with aqueous phase analysis and the inner droplet environment cannot easily communicate with the external phase. We present a high-throughput, miniaturized screening platform for microchip-synthesized genes using microfluidics-generated water-in-oil-in-water (W/O/W) double emulsion (DE) droplets that overcome these limitations. Synthetic gene variants of fluorescent proteins are synthesized with a custom-built microarray inkjet synthesizer, which are then screened for expression in Escherichia coli (E. coli) cells. Bacteria bearing individual fluorescent gene variants are encapsulated as single cells into DE droplets where fluorescence signals are enhanced by 100 times within 24 h of proliferation. Enrichment of functionally-correct genes by employing an error correction method is demonstrated by screening DE droplets containing fluorescent clones of bacteria with the red fluorescent protein (rfp) gene. Permeation of isopropyl β-D-1-thiogalactopyranoside (IPTG) through the thin oil layer from the external solution initiates target gene expression. The induced expression of the synthetic fluorescent proteins from at least ∼100 bacteria per droplet generates detectable fluorescence signals to enable fluorescence-activated cell sorting (FACS) of the intact droplets. This technology obviates time- and labor-intensive cell culture typically required in conventional bulk experiment.


 Please wait while we load your content...
Please wait while we load your content...