A systematic reconstruction and constraint-based analysis of Leishmania donovani metabolic network: identification of potential antileishmanial drug targets†
Abstract
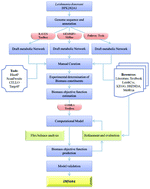
Visceral leishmaniasis, a lethal parasitic disease, is caused by the protozoan parasite Leishmania donovani. The absence of an effective vaccine, drug toxicity and parasite resistance necessitates the identification of novel drug targets. Reconstruction of genome-scale metabolic models and their simulation has been established as an important tool for systems-level understanding of a microorganism's metabolism. In this work, amalgamating the tools and techniques of computational systems biology with rigorous manual curation, a constraint-based metabolic model for Leishmania donovani BPK282A1 has been developed. New functional annotations for 18 formerly hypothetical or erroneously annotated genes (encountered during iterative refinement of the model) have been proposed. Further, to formulate an accurate biomass objective function, experimental determination of previously uncharacterized biomass constituents was performed. The developed model is a highly compartmentalized metabolic model, comprising 1159 reactions, 1135 metabolites and 604 genes. The model exhibited around 76% accuracy for the prediction of experimental phenotypes of gene knockout studies and drug inhibition assays. Employing in silico gene knockout studies, we identified 28 essential genes with negligible sequence identity to the human proteins. Moreover, by dissecting the functional interdependencies of metabolic pathways, 70 synthetic lethal pairs were identified. Finally, in order to delineate stage-specific metabolism, gene-expression data of the amastigote stage residing in human macrophages were integrated into the model. By comparing the flux distribution, we illustrated the stage-specific differences in metabolism and environmental conditions that are in good agreement with the experimental findings. The developed model can serve as a highly enriched knowledgebase of legacy data and an important tool for generating experimentally verifiable hypotheses.


 Please wait while we load your content...
Please wait while we load your content...