Investigating d-lysine stereochemistry for epigenetic methylation, demethylation and recognition†
Abstract
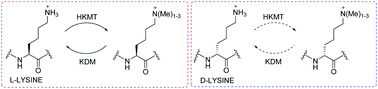
Histone lysine methylation is regulated by Nε-methyltransferases, demethylases, and Nε-methyl lysine binding proteins. Thermodynamic, catalytic and computational studies were carried out to investigate the interaction of three epigenetic protein classes with synthetic histone substrates containing L- and D-lysine residues. The results reveal that out of the three classes, Nε-methyl lysine binding proteins are superior in accepting lysines with the D-configuration.


 Please wait while we load your content...
Please wait while we load your content...