Gene expression profiling to identify the toxicities and potentially relevant disease outcomes due to endosulfan exposure†
Abstract
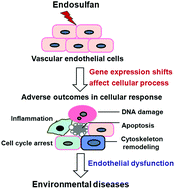
Endosulfan, one of the most toxic organochlorine pesticides, belongs to a group of persistent organic pollutants. Gene expression profiling offers a promising approach in health hazard identification of chemicals. The aim of this study was to use gene expression profiling to identify the toxicities and potentially relevant human diseases due to endosulfan exposure. We performed DNA microarray analysis to analyze gene expression profiles in human endothelial cells exposed to 20, 40 and 60 μM endosulfan in combination with an endothelial phenotype. Microarray results showed that endosulfan increased the number of altered genes in a dose-dependent manner, and changed the expression of 161 genes across all treatment groups. qRT-PCR closely matched the microarray data for the genes tested. Significantly enriched biological processes for overlapping down-regulated genes include the neurological system process, signal transduction, and homeostatic process in all the dose groups. These down-regulated genes were associated with cytoskeleton organization and DNA repair at low doses, and involved in cell cycle, apoptosis, p53 pathway and carcinogenesis at high doses. Those up-regulated genes were linked to the inflammatory response and transcriptional misregulation in cancer at higher doses. These findings are consistent with our established endothelial phenotypes. Endosulfan may be relevant to human diseases including liver cancer, prostate cancer and leukemia using the NextBio Human Disease Atlas. These results provide molecular evidence supporting the toxicities and carcinogenic potential of endosulfan in humans.

 Please wait while we load your content...
Please wait while we load your content...