Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus†
Abstract
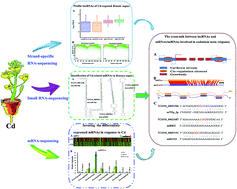
There is increasing evidence of long non-coding RNA (lncRNA) involvement in a variety of biological responses to environmental stresses. The toxic heavy metal cadmium (Cd) is one of the major inorganic contaminants in environments. How lncRNAs are involved in Cd stress-induced response and regulatory mechanism in dicot plants is unknown. The present study performed strand-specific RNA-sequencing (RNA-Seq) to profile lncRNAs in Cd-exposed rapeseed (Brassica napus). More than 60 million clean reads corresponding to 5038 unique B. napus lncRNAs were identified, in which 2546 lncRNAs were generated from the sense strand and 2492 were generated from natural antisense transcripts (lncNATs). A combinational analysis resulted in identification of 301 lncRNAs responding to Cd stress. Cd-induced expression of lncRNAs appeared to be associated with their nearby protein-coding genes. Many coding genes flanking lncRNA loci were involved in Cd uptake, translocation and stress-responses, suggesting that the associated lncRNAs might have similar functions. Four lncRNAs were identified as precursors of miR824, miR167d, miR156d and miR156e. Sixty-seven lncRNAs acted as competing endogenous target mimics (eTMs) for 36 miRNAs potentially involved in Cd stress response. The functional mimics were validated by transient transformation of three lncRNAs into B. napus protoplasts under control of double 35S promoter with a decoy lncRNA (d35S-lncRNAs-pAN580). These results suggest that a set of putative lncRNAs were involved in response to Cd stress in B. napus.

 Please wait while we load your content...
Please wait while we load your content...