Programmable DNA scaffolds for spatially-ordered protein assembly†
Abstract
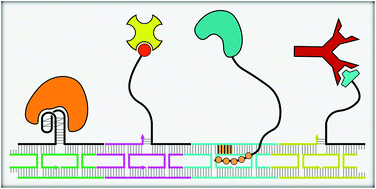
Ever since the notion of using DNA as a material was realized, it has been employed in the construction of complex structures that facilitate the assembly of nanoparticles or macromolecules with nanometer-scale precision. Specifically, tiles fashioned from DNA strands and DNA origami sheets have been shown to be suitable as scaffolds for immobilizing proteins with excellent control over their spatial positioning. Supramolecular assembly of proteins into periodic arrays in one or more dimensions is one of the most challenging aspects in the design of scaffolds for biomolecular investigations and macromolecular crystallization. This review provides a brief overview of how various biomolecular interactions with high degree of specificity such as streptavidin–biotin, antigen–antibody, and aptamer–protein interactions have been used to fabricate linear and multidimensional assemblies of structurally intact and functional proteins. The use of DNA-binding proteins as adaptors, polyamide recognition on DNA scaffolds and oligonucleotide linkers for protein assembly are also discussed.

 Please wait while we load your content...
Please wait while we load your content...